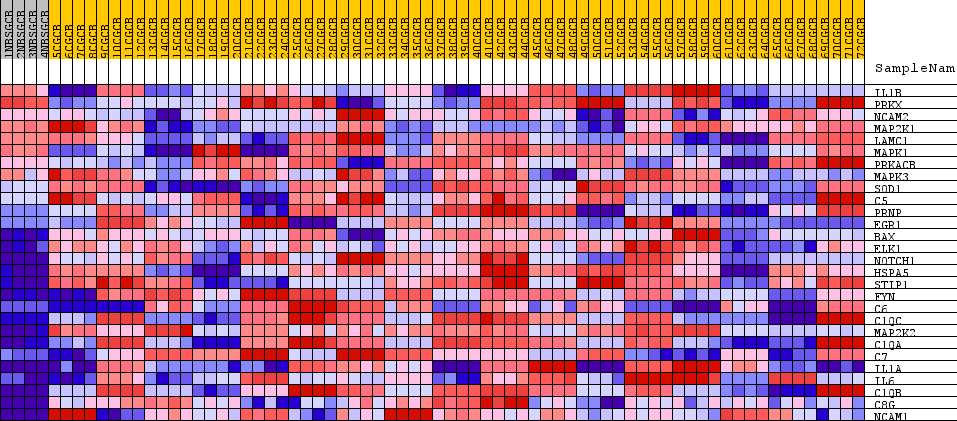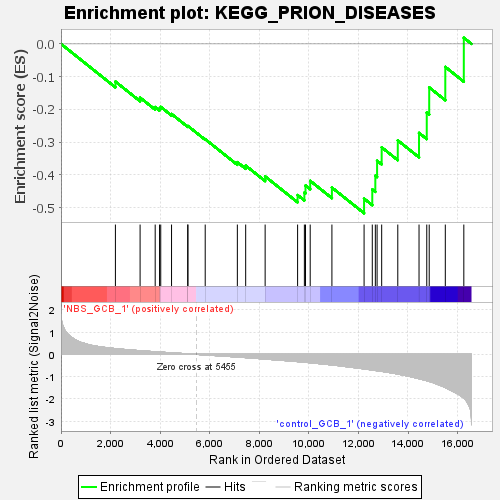
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GCB_NBS_vs_control.NBS_GCB_1_vs_control_GCB_1.cls #NBS_GCB_1_versus_control_GCB_1 |
| Phenotype | NBS_GCB_1_vs_control_GCB_1.cls#NBS_GCB_1_versus_control_GCB_1 |
| Upregulated in class | control_GCB_1 |
| GeneSet | KEGG_PRION_DISEASES |
| Enrichment Score (ES) | -0.5174269 |
| Normalized Enrichment Score (NES) | -1.2764734 |
| Nominal p-value | 0.11574557 |
| FDR q-value | 0.8704204 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | IL1B | NA | 2195 | 0.258 | -0.1151 | No |
| 2 | PRKX | NA | 3191 | 0.163 | -0.1642 | No |
| 3 | NCAM2 | NA | 3800 | 0.117 | -0.1930 | No |
| 4 | MAP2K1 | NA | 3976 | 0.103 | -0.1966 | No |
| 5 | LAMC1 | NA | 4024 | 0.100 | -0.1926 | No |
| 6 | MAPK1 | NA | 4460 | 0.069 | -0.2142 | No |
| 7 | PRKACB | NA | 5113 | 0.024 | -0.2519 | No |
| 8 | MAPK3 | NA | 5121 | 0.023 | -0.2508 | No |
| 9 | SOD1 | NA | 5818 | -0.026 | -0.2911 | No |
| 10 | C5 | NA | 7115 | -0.118 | -0.3614 | No |
| 11 | PRNP | NA | 7451 | -0.145 | -0.3718 | No |
| 12 | EGR1 | NA | 8236 | -0.209 | -0.4049 | No |
| 13 | BAX | NA | 9546 | -0.324 | -0.4619 | No |
| 14 | ELK1 | NA | 9819 | -0.352 | -0.4544 | No |
| 15 | NOTCH1 | NA | 9865 | -0.357 | -0.4327 | No |
| 16 | HSPA5 | NA | 10054 | -0.375 | -0.4184 | No |
| 17 | STIP1 | NA | 10930 | -0.474 | -0.4390 | No |
| 18 | FYN | NA | 12228 | -0.653 | -0.4728 | Yes |
| 19 | C6 | NA | 12559 | -0.702 | -0.4448 | Yes |
| 20 | C1QC | NA | 12682 | -0.724 | -0.4028 | Yes |
| 21 | MAP2K2 | NA | 12753 | -0.733 | -0.3570 | Yes |
| 22 | C1QA | NA | 12938 | -0.762 | -0.3161 | Yes |
| 23 | C7 | NA | 13588 | -0.885 | -0.2949 | Yes |
| 24 | IL1A | NA | 14445 | -1.093 | -0.2720 | Yes |
| 25 | IL6 | NA | 14757 | -1.184 | -0.2099 | Yes |
| 26 | C1QB | NA | 14856 | -1.213 | -0.1330 | Yes |
| 27 | C8G | NA | 15505 | -1.491 | -0.0704 | Yes |
| 28 | NCAM1 | NA | 16252 | -1.964 | 0.0187 | Yes |