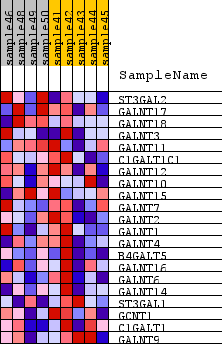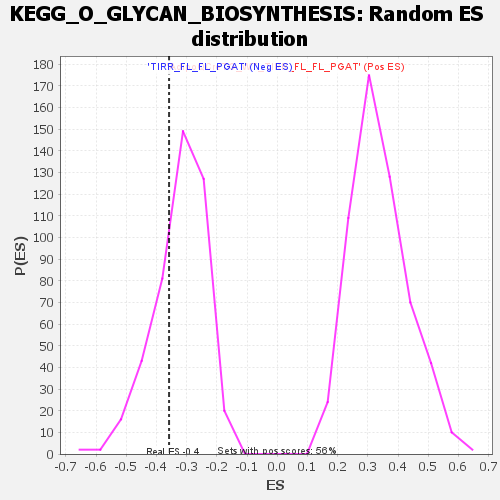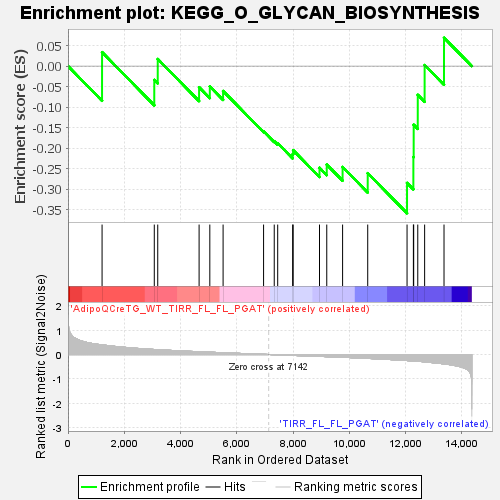
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
The same image in compressed SVG format
| Dataset | TIRRKO_WT_specific_PGAT.AdipoQCreTG_WT_TIRR_FL_FL_PGAT_vs_TIRR_FL_FL_PGAT.cls #AdipoQCreTG_WT_TIRR_FL_FL_PGAT_versus_TIRR_FL_FL_PGAT.AdipoQCreTG_WT_TIRR_FL_FL_PGAT_vs_TIRR_FL_FL_PGAT.cls #AdipoQCreTG_WT_TIRR_FL_FL_PGAT_versus_TIRR_FL_FL_PGAT_repos |
| Phenotype | AdipoQCreTG_WT_TIRR_FL_FL_PGAT_vs_TIRR_FL_FL_PGAT.cls#AdipoQCreTG_WT_TIRR_FL_FL_PGAT_versus_TIRR_FL_FL_PGAT_repos |
| Upregulated in class | TIRR_FL_FL_PGAT |
| GeneSet | KEGG_O_GLYCAN_BIOSYNTHESIS |
| Enrichment Score (ES) | -0.35818252 |
| Normalized Enrichment Score (NES) | -1.1127528 |
| Nominal p-value | 0.275 |
| FDR q-value | 0.5559556 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ST3GAL2 | n/a | 1211 | 0.394 | 0.0339 | No |
| 2 | GALNT17 | n/a | 3067 | 0.206 | -0.0337 | No |
| 3 | GALNT18 | n/a | 3190 | 0.198 | 0.0172 | No |
| 4 | GALNT3 | n/a | 4660 | 0.112 | -0.0515 | No |
| 5 | GALNT11 | n/a | 5041 | 0.095 | -0.0495 | No |
| 6 | C1GALT1C1 | n/a | 5513 | 0.072 | -0.0606 | No |
| 7 | GALNT12 | n/a | 6952 | 0.008 | -0.1586 | No |
| 8 | GALNT10 | n/a | 7331 | -0.007 | -0.1829 | No |
| 9 | GALNT15 | n/a | 7454 | -0.012 | -0.1877 | No |
| 10 | GALNT7 | n/a | 7986 | -0.035 | -0.2143 | No |
| 11 | GALNT2 | n/a | 8003 | -0.036 | -0.2047 | No |
| 12 | GALNT1 | n/a | 8940 | -0.075 | -0.2476 | No |
| 13 | GALNT4 | n/a | 9198 | -0.086 | -0.2398 | No |
| 14 | B4GALT5 | n/a | 9761 | -0.111 | -0.2456 | No |
| 15 | GALNT16 | n/a | 10654 | -0.157 | -0.2607 | No |
| 16 | GALNT6 | n/a | 12052 | -0.246 | -0.2844 | Yes |
| 17 | GALNT14 | n/a | 12277 | -0.262 | -0.2213 | Yes |
| 18 | ST3GAL1 | n/a | 12288 | -0.264 | -0.1428 | Yes |
| 19 | GCNT1 | n/a | 12433 | -0.276 | -0.0700 | Yes |
| 20 | C1GALT1 | n/a | 12677 | -0.298 | 0.0025 | Yes |
| 21 | GALNT9 | n/a | 13366 | -0.381 | 0.0689 | Yes |