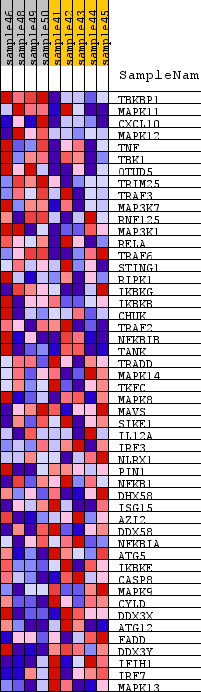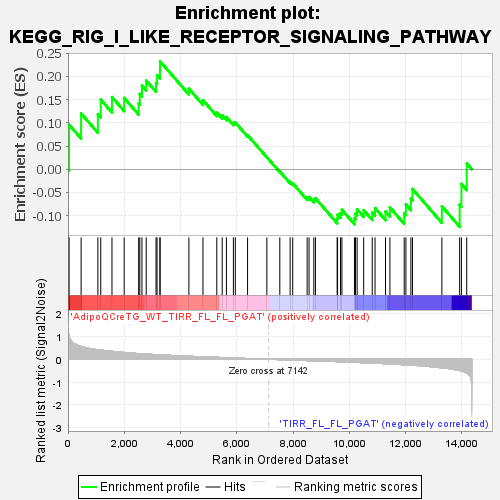
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
The same image in compressed SVG format
| Dataset | TIRRKO_WT_specific_PGAT.AdipoQCreTG_WT_TIRR_FL_FL_PGAT_vs_TIRR_FL_FL_PGAT.cls #AdipoQCreTG_WT_TIRR_FL_FL_PGAT_versus_TIRR_FL_FL_PGAT.AdipoQCreTG_WT_TIRR_FL_FL_PGAT_vs_TIRR_FL_FL_PGAT.cls #AdipoQCreTG_WT_TIRR_FL_FL_PGAT_versus_TIRR_FL_FL_PGAT_repos |
| Phenotype | AdipoQCreTG_WT_TIRR_FL_FL_PGAT_vs_TIRR_FL_FL_PGAT.cls#AdipoQCreTG_WT_TIRR_FL_FL_PGAT_versus_TIRR_FL_FL_PGAT_repos |
| Upregulated in class | AdipoQCreTG_WT_TIRR_FL_FL_PGAT |
| GeneSet | KEGG_RIG_I_LIKE_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | 0.23247108 |
| Normalized Enrichment Score (NES) | 0.8331386 |
| Nominal p-value | 0.7602627 |
| FDR q-value | 0.9178983 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | TBKBP1 | n/a | 39 | 1.028 | 0.0960 | Yes |
| 2 | MAPK11 | n/a | 466 | 0.561 | 0.1201 | Yes |
| 3 | CXCL10 | n/a | 1062 | 0.417 | 0.1186 | Yes |
| 4 | MAPK12 | n/a | 1164 | 0.401 | 0.1500 | Yes |
| 5 | TNF | n/a | 1563 | 0.346 | 0.1554 | Yes |
| 6 | TBK1 | n/a | 1997 | 0.295 | 0.1535 | Yes |
| 7 | OTUD5 | n/a | 2512 | 0.247 | 0.1414 | Yes |
| 8 | TRIM25 | n/a | 2552 | 0.244 | 0.1621 | Yes |
| 9 | TRAF3 | n/a | 2629 | 0.239 | 0.1797 | Yes |
| 10 | MAP3K7 | n/a | 2780 | 0.226 | 0.1910 | Yes |
| 11 | RNF125 | n/a | 3130 | 0.202 | 0.1860 | Yes |
| 12 | MAP3K1 | n/a | 3165 | 0.200 | 0.2028 | Yes |
| 13 | RELA | n/a | 3269 | 0.193 | 0.2141 | Yes |
| 14 | TRAF6 | n/a | 3272 | 0.193 | 0.2325 | Yes |
| 15 | STING1 | n/a | 4297 | 0.131 | 0.1735 | No |
| 16 | RIPK1 | n/a | 4798 | 0.106 | 0.1488 | No |
| 17 | IKBKG | n/a | 5289 | 0.083 | 0.1225 | No |
| 18 | IKBKB | n/a | 5481 | 0.074 | 0.1162 | No |
| 19 | CHUK | n/a | 5631 | 0.067 | 0.1123 | No |
| 20 | TRAF2 | n/a | 5879 | 0.056 | 0.1004 | No |
| 21 | NFKBIB | n/a | 5948 | 0.053 | 0.1007 | No |
| 22 | TANK | n/a | 6383 | 0.032 | 0.0735 | No |
| 23 | TRADD | n/a | 7068 | 0.003 | 0.0260 | No |
| 24 | MAPK14 | n/a | 7529 | -0.016 | -0.0046 | No |
| 25 | TKFC | n/a | 7895 | -0.031 | -0.0272 | No |
| 26 | MAPK8 | n/a | 7988 | -0.035 | -0.0303 | No |
| 27 | MAVS | n/a | 8500 | -0.056 | -0.0606 | No |
| 28 | SIKE1 | n/a | 8569 | -0.059 | -0.0596 | No |
| 29 | IL12A | n/a | 8738 | -0.067 | -0.0650 | No |
| 30 | IRF3 | n/a | 8795 | -0.069 | -0.0623 | No |
| 31 | NLRX1 | n/a | 9567 | -0.102 | -0.1064 | No |
| 32 | PIN1 | n/a | 9582 | -0.103 | -0.0975 | No |
| 33 | NFKB1 | n/a | 9686 | -0.108 | -0.0944 | No |
| 34 | DHX58 | n/a | 9736 | -0.110 | -0.0872 | No |
| 35 | ISG15 | n/a | 10182 | -0.132 | -0.1056 | No |
| 36 | AZI2 | n/a | 10219 | -0.134 | -0.0953 | No |
| 37 | DDX58 | n/a | 10278 | -0.138 | -0.0861 | No |
| 38 | NFKBIA | n/a | 10510 | -0.149 | -0.0879 | No |
| 39 | ATG5 | n/a | 10815 | -0.167 | -0.0932 | No |
| 40 | IKBKE | n/a | 10914 | -0.172 | -0.0836 | No |
| 41 | CASP8 | n/a | 11286 | -0.193 | -0.0909 | No |
| 42 | MAPK9 | n/a | 11443 | -0.204 | -0.0822 | No |
| 43 | CYLD | n/a | 11950 | -0.238 | -0.0948 | No |
| 44 | DDX3X | n/a | 12011 | -0.242 | -0.0757 | No |
| 45 | ATG12 | n/a | 12188 | -0.256 | -0.0634 | No |
| 46 | FADD | n/a | 12245 | -0.260 | -0.0424 | No |
| 47 | DDX3Y | n/a | 13290 | -0.372 | -0.0796 | No |
| 48 | IFIH1 | n/a | 13922 | -0.492 | -0.0765 | No |
| 49 | IRF7 | n/a | 13984 | -0.511 | -0.0316 | No |
| 50 | MAPK13 | n/a | 14173 | -0.598 | 0.0127 | No |